import rpy25 Interfacing R from Python
While Python is quite versatile language that makes solving problems very simple, the language R has its own strengths. It has been very popular in the academic circles and many of the bioinformatics algorithms often gets released as R packages. Often, the only possible way solve a particular problem effectively is using R.
It is pragmatic to use the right tool for the right job. In this session, we’ll see how to use R from Python. We’ll see two different approaches - using R from jupyter notebooks along with Python and using R in scripts and streamlit applications.
5.1 The rpy2 package
The rpy2 package in Python provides a way to execute R from python and it also has good support for jupyter notebooks.
5.1.1 Using R in the notebook
Import rpy2 package.
And load rpy2 jupyter extension.
%load_ext rpy2.ipythonNow we can run R code and even plot graphs.
%%R
library(palmerpenguins)
# Print the first 6 rows
head(penguins, 6)
plot(penguins$bill_length_mm, penguins$bill_depth_mm)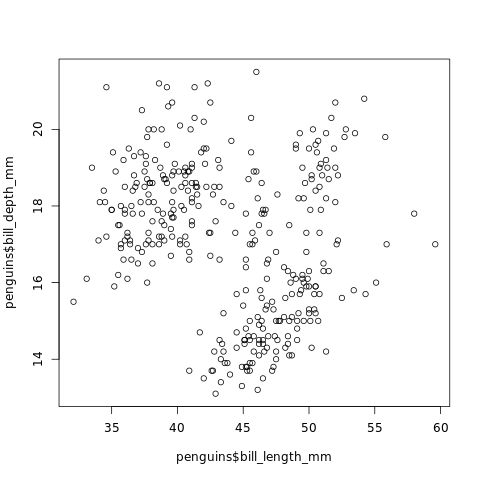
You can even use the familiar ggplot2 library.
%%R
# Whatever we have run earlier in R will still be available here.
library(ggplot2)
ggplot(penguins, aes(bill_length_mm, bill_depth_mm, colour = species)) +
geom_point()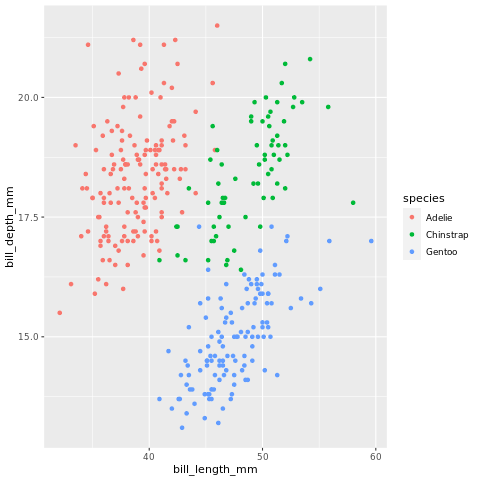
5.2 Passing values between Python and R
import pandas as pdDefine two of values in Python.
x = 10
y = 20Add add them in R.
%%R -i x -i y
x + y[1] 30It is even possible to access the value of a variable define in R from Python, but we’ll look into into it later.
Interesting part of this approach is that we can even pass dataframes into R.
df = pd.read_csv("../shared/un-min.csv")df.head()| country | region | lifeMale | lifeFemale | infantMortality | GDPperCapita | |
|---|---|---|---|---|---|---|
| 0 | Afghanistan | Asia | 45.0 | 46.0 | 154 | 2848 |
| 1 | Albania | Europe | 68.0 | 74.0 | 32 | 863 |
| 2 | Algeria | Africa | 67.5 | 70.3 | 44 | 1531 |
| 3 | Angola | Africa | 44.9 | 48.1 | 124 | 355 |
| 4 | Argentina | America | 69.6 | 76.8 | 22 | 8055 |
%%R -i df
library(ggplot2)
ggplot(df, aes(GDPperCapita, infantMortality, colour = region)) +
geom_point()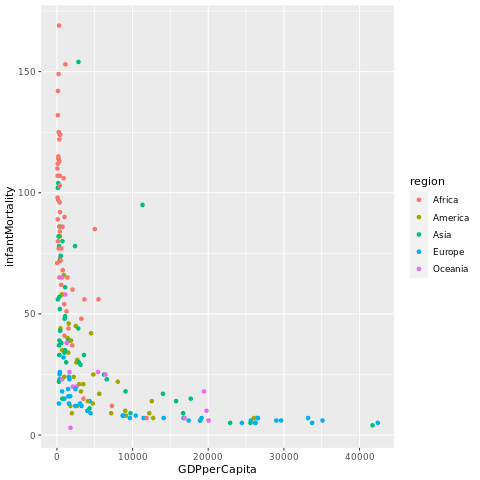
Question: Can we read dataframe in R and get it to Python and plot?
An easy way is to write the dataframe in R to a file and read it in python. We’ll look into another approach in a later section.
%%R
data(mtcars)
head(mtcars)
write.csv(mtcars, "mtcars.csv")!ls -l mtcars.csv-rw-r--r-- 1 anand anand 1783 Jun 3 03:59 mtcars.csvdf = pd.read_csv("mtcars.csv")df.head()| Unnamed: 0 | mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear | carb | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Mazda RX4 | 21.0 | 6 | 160.0 | 110 | 3.90 | 2.620 | 16.46 | 0 | 1 | 4 | 4 |
| 1 | Mazda RX4 Wag | 21.0 | 6 | 160.0 | 110 | 3.90 | 2.875 | 17.02 | 0 | 1 | 4 | 4 |
| 2 | Datsun 710 | 22.8 | 4 | 108.0 | 93 | 3.85 | 2.320 | 18.61 | 1 | 1 | 4 | 1 |
| 3 | Hornet 4 Drive | 21.4 | 6 | 258.0 | 110 | 3.08 | 3.215 | 19.44 | 1 | 0 | 3 | 1 |
| 4 | Hornet Sportabout | 18.7 | 8 | 360.0 | 175 | 3.15 | 3.440 | 17.02 | 0 | 0 | 3 | 2 |
df.plot(kind="scatter", x="mpg", y="hp")<Axes: xlabel='mpg', ylabel='hp'>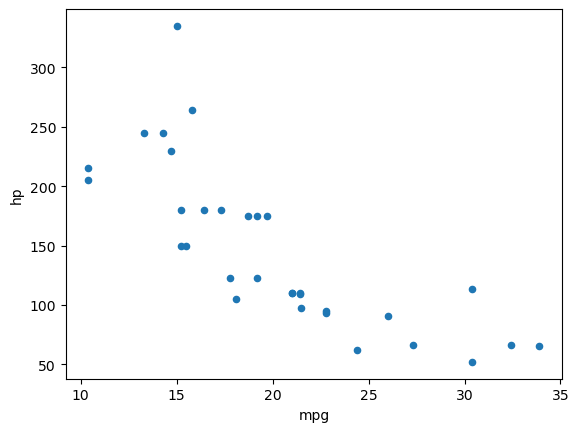
Problem: Plot a graph of lifeMale vs. lifeFemale, color by region and size by th GDP using ggplot2 in R.
Problem: Read iris dataset from ../shared/iris.csv in Python and plot a scatter plot on SepalLength vs. SepalWidth and color by Name using R.
5.3 Invoking R programatically
Invoking R programatically can be done using the rpy2 package without using the %%R magic method.
The rpy2 module allows passing inputs to R and read the results back. While there may be issues with convertion of types between python and R, it will be simpler if we stick to primitive and well known types.
To make things simple for you, I have created a function rexec.
import rpy2.robjects as robjects
from rpy2.robjects import pandas2ri
from rpy2.robjects.conversion import localconverter
def rexec(code, **params):
"""Script to run R code passing some inputs to it.
>>> rexec("print(x+y)", x=10, y=20)
[1] 30
If you would like to get back some result from R, assign that
to variable result.
>>> rexec("result <- x + y", x=10, y=20)
array([30], dtype=int32)
"""
# rpy2 has some multi-threading issues and this is the recommended way to convert python objects to R
# See https://github.com/rpy2/rpy2/issues/978
with localconverter(robjects.default_converter + pandas2ri.converter) as cv:
robjects.globalenv['result'] = False
for k, v in params.items():
robjects.globalenv[k] = cv.py2rpy(v)
robjects.r(code)
result = robjects.r.get("result")
if result is not None:
return cv.rpy2py(result)rexec("print(x+y)", x=10, y=20)[1] 30| 0 |
z = rexec("result <- x+y", x=10, y=20)zarray([30], dtype=int32)z[0]305.3.1 Ploting and Saving Graphs
Now, let us look at running a complete R script from python by passing inputs and outputs only through files.
It reads the input from ../shared/un-min.csv and write the graph as image to rplot.png.
df = pd.read_csv("../shared/un-min.csv")df.head()| country | region | lifeMale | lifeFemale | infantMortality | GDPperCapita | |
|---|---|---|---|---|---|---|
| 0 | Afghanistan | Asia | 45.0 | 46.0 | 154 | 2848 |
| 1 | Albania | Europe | 68.0 | 74.0 | 32 | 863 |
| 2 | Algeria | Africa | 67.5 | 70.3 | 44 | 1531 |
| 3 | Angola | Africa | 44.9 | 48.1 | 124 | 355 |
| 4 | Argentina | America | 69.6 | 76.8 | 22 | 8055 |
code = """
library(ggplot2)
ggplot(df, aes(GDPperCapita, lifeFemale, colour = region)) +
geom_point()
ggsave(filename = image_path)
"""rexec(code, df=df, image_path="rplot.png")Saving 7 x 7 in image| 0 |
!ls -l *.png-rw-r--r-- 1 anand anand 136075 Jun 3 04:13 rplot.pngfrom IPython.display import ImageImage(filename="rplot.png")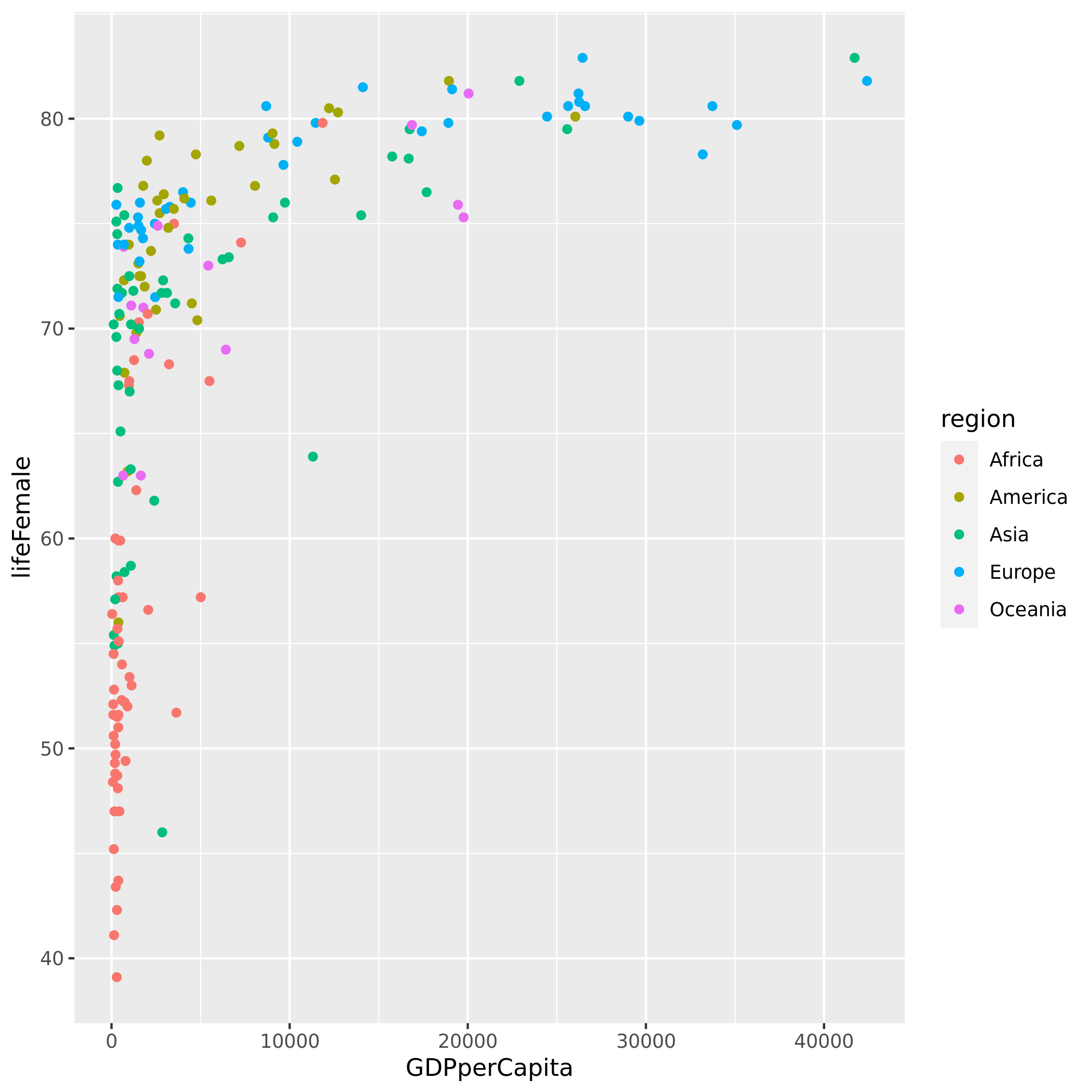
Q: Can we do some analysis in R and get the results back in Python for plotting?
Let’s take iris dataset, pass it to R for aggregation, get the result back to Python and plot them.
df.head()| country | region | lifeMale | lifeFemale | infantMortality | GDPperCapita | |
|---|---|---|---|---|---|---|
| 0 | Afghanistan | Asia | 45.0 | 46.0 | 154 | 2848 |
| 1 | Albania | Europe | 68.0 | 74.0 | 32 | 863 |
| 2 | Algeria | Africa | 67.5 | 70.3 | 44 | 1531 |
| 3 | Angola | Africa | 44.9 | 48.1 | 124 | 355 |
| 4 | Argentina | America | 69.6 | 76.8 | 22 | 8055 |
df.set_index("country", inplace=True)df.head()| region | lifeMale | lifeFemale | infantMortality | GDPperCapita | |
|---|---|---|---|---|---|
| country | |||||
| Afghanistan | Asia | 45.0 | 46.0 | 154 | 2848 |
| Albania | Europe | 68.0 | 74.0 | 32 | 863 |
| Algeria | Africa | 67.5 | 70.3 | 44 | 1531 |
| Angola | Africa | 44.9 | 48.1 | 124 | 355 |
| Argentina | America | 69.6 | 76.8 | 22 | 8055 |
code = """
df2 <- aggregate(. ~ region, df, mean)
result <- df2
"""rexec(code, df=df)| region | lifeMale | lifeFemale | infantMortality | GDPperCapita | |
|---|---|---|---|---|---|
| 1 | Africa | 52.052830 | 55.286792 | 86.320755 | 1217.641509 |
| 2 | America | 69.082857 | 74.474286 | 26.657143 | 5080.085714 |
| 3 | Asia | 65.373913 | 69.439130 | 43.782609 | 5453.195652 |
| 4 | Europe | 70.362500 | 77.545000 | 11.575000 | 12860.050000 |
| 5 | Oceania | 67.464286 | 72.092857 | 24.642857 | 7131.785714 |